Genomic Comparison Study:
- HSV-2 to HSV-1 genome comparison
HSV-2 and HSV-1 share the same overall structure, as well as very high similarity in most genes. The G+C content of the HSV-2 genome is 70.4%, compared with 68.3% for HSV-1. Strains of HSV-1 differ from the strains of HSV-2 at most of their cleavage sites. Only half of the HSV-1 genome is complementary to the HSV-2 genome and that in the homologous regions there is about 15% mismatch in base pairing as judged by the melting temperature of the hybrid. This means that in the homologous regions, one in seven bases differs between HSV-1 and HSV-2.
The BugSpray comparison below shows genome alignment based on similar genes and dramatically illustrates the similarity of gene arrangement between the HSV-1 and HSV-2 genomes. BugSpray draws lines from genes on the top genome, in this case HSV-2, to the gene with the best BLAST hit in the bottom genome, here HSV-1. Green lines indicate pairs of genes that code on the same strand and red lines indicate gene pairs that code on the opposite strand. Clearly there are no rearrangements or reordering of genes between these two viruses. The three red lines show best hits of gamma 34.5, alpha4 and alpha0 to the copy in the inverted repeat, because BugSpray arbitrarily selects only one best hit if there are multiple hits with the same weight. Only Orf-P does not have a BLASTp hit to a gene in the HSV-1 genome.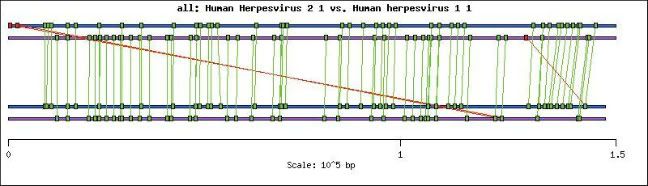
Figure: BUGSPRAY GENOME ALIGNMENT. Nina Thayer, 2001: HSV-2 Genome and Proteome Properties.
- HSV-2 to SA8 genome comparison
Herpesvirus simian agent 8 (SA8; cercopithecine herpesvirus 2) is a baboon simplexvirus closely related to HSV-2. The genome of SA8 is 150,715 bp long, with an overall G+C content of 76%, the highest among the simplexviruses sequenced so far, compared to HSV-2 in length of 154, 746 bp with base composition of 70.4% G+C content. The genomic arrangement of SA8 is similar to that of other simplexviruses, that is unique long (UL) and unique short (US) regions bordered by 2 sets of inverted repeats, while HSV-2 has 3 sets of inverted repeats.
References:
1. Nina Thayer, 2001: Human Herpesvirus 2 (HSV-2). Internet: http://stdgen.northwestern.edu/stdgen/bacteria/hhv2/hhv2properties.html.
2. American Society for Microbiology, April 1975: Structure and Function of Herpesvirus Genomes. Internet: http://jvi.asm.org/cgi/reprint/15/4/726.pdf
3. Shaun D. Tyler et al., 2004: Complete genome sequence of cercopithecine herpesvirus 2 (SA8) and comparison with other simplexviruses. Internet: http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WXR-4DSR13J-2&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=f122e9d1acdb89157dc6240f4e6902c0
Comments and inquiries are always welcome.
Prepared by Tan Phit Ling 1071114866

No comments:
Post a Comment